Rows: 2,000
Columns: 10
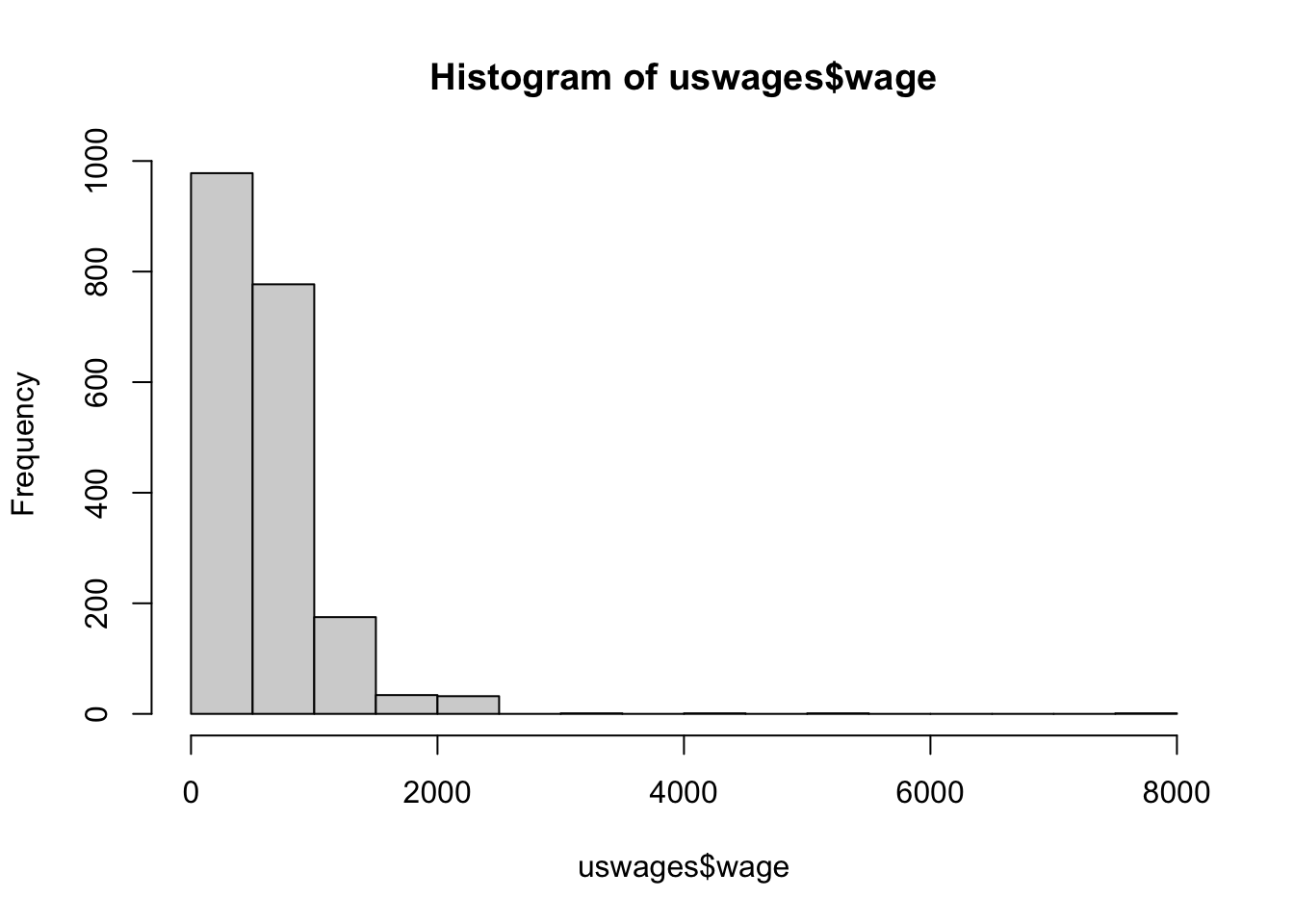
$ wage <dbl> 771.60, 617.28, 957.83, 617.28, 902.18, 299.15, 541.31, 148.39, …
$ educ <int> 18, 15, 16, 12, 14, 12, 16, 16, 12, 12, 9, 14, 17, 14, 14, 10, 1…
$ exper <int> 18, 20, 9, 24, 12, 33, 42, 0, 36, 37, 20, 29, 16, 21, 11, 10, 8,…
$ race <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ smsa <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 0, 1, 0, 1, 0, 1, 1, 1, 0, 1…
$ ne <int> 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0…
$ mw <int> 0, 0, 0, 0, 1, 0, 0, 1, 0, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0…
$ so <int> 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 1, 1, 0, 0, 0, 0, 1, 1, 0, 1, 1, 1…
$ we <int> 0, 1, 0, 0, 0, 1, 0, 0, 1, 0, 0, 0, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0…
$ pt <int> 0, 0, 0, 0, 0, 0, 1, 1, 1, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0…